Multicellular organisms, including humans, are composed of various organs, tissues, and cell types. Although all cells share the same genome, what causes this diversity? Moreover, why do cells arranged in three-dimensional configurations exhibit unique functions based on their spatial locations?
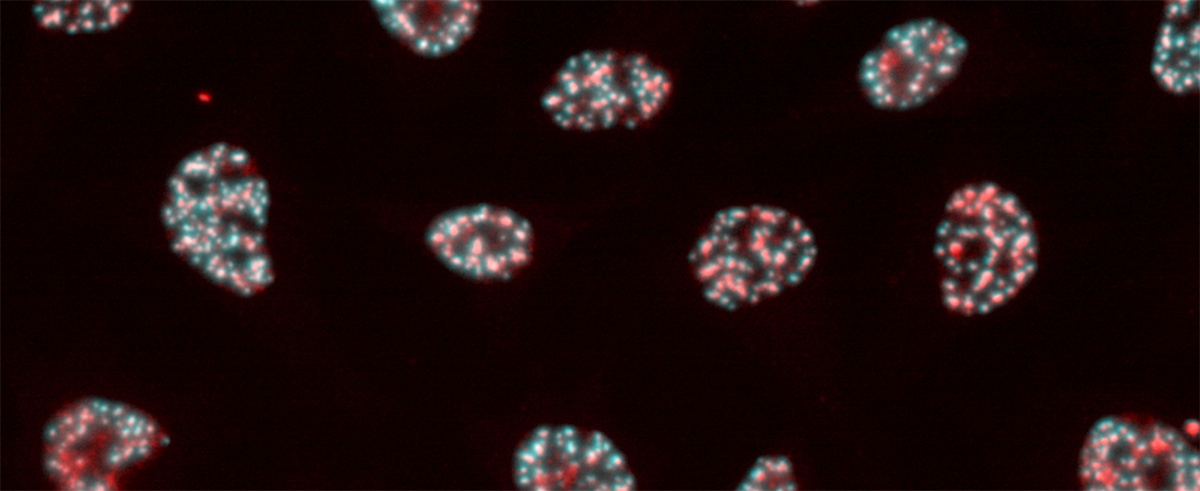
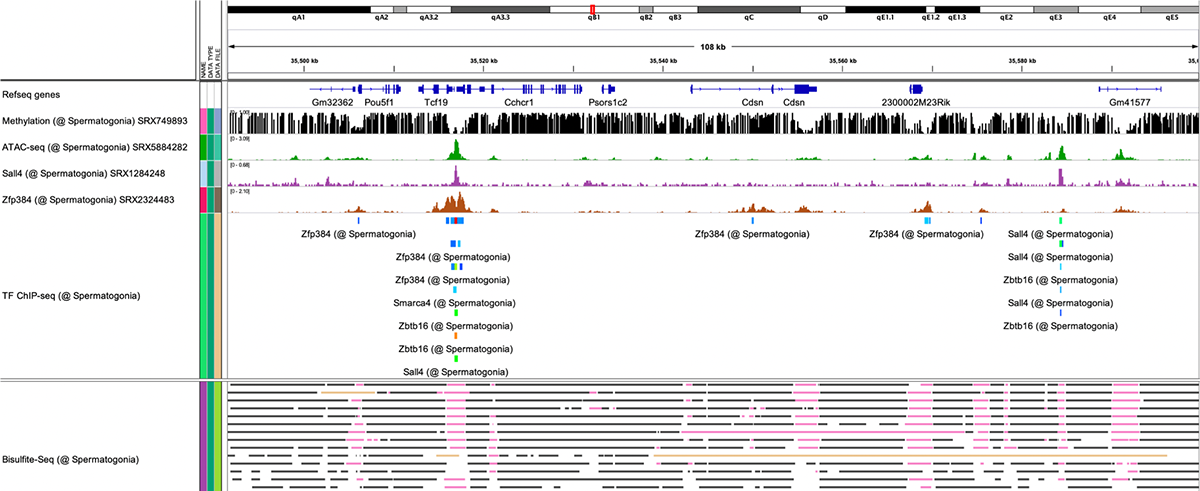
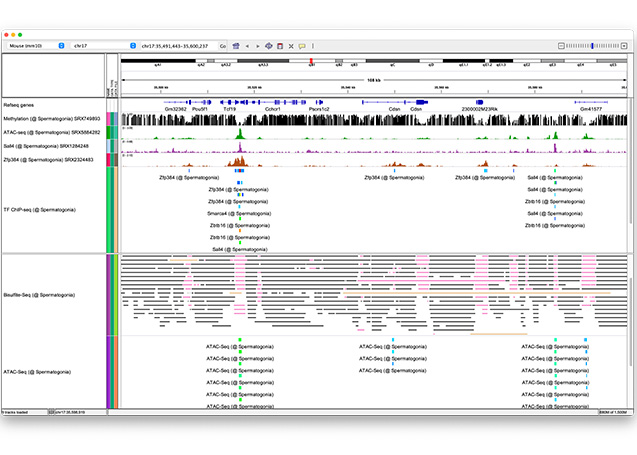
The solution to this mystery lies in spatiotemporal gene expression. The Oki Lab has developed a technique called PIC to comprehensively elucidate genes expressed locally. In addition, ChIP-Atlas, a data-mining suite for exploring epigenomic landscapes, has been developed to unveil the regulatory mechanisms of gene expression. By integrating these two approaches in our research, we are aiming to elucidate the etiology of diseases and advance applications in drug discovery.
News
- 2025-02-28沖 真弥さん、鄒 兆南さんが京都大学で開催された企業向けセミナー「脱 Dry!脱 写経!ゲノムインフォマティクス演習」にて講師を務めました。
- 2025-02-05沖 真弥、鄒 兆南さんが埼玉県立がんセンター臨床腫瘍研究所で ChIP-Atlas ハンズオンセミナーを開催しました。
- 2025-01-30沖 真弥さんが第7回 ExCELLS シンポジウムで講演しました。
- 2025-01-29沖 真弥さんが熊本大学 健康長寿代謝制御研究センターで講演しました。
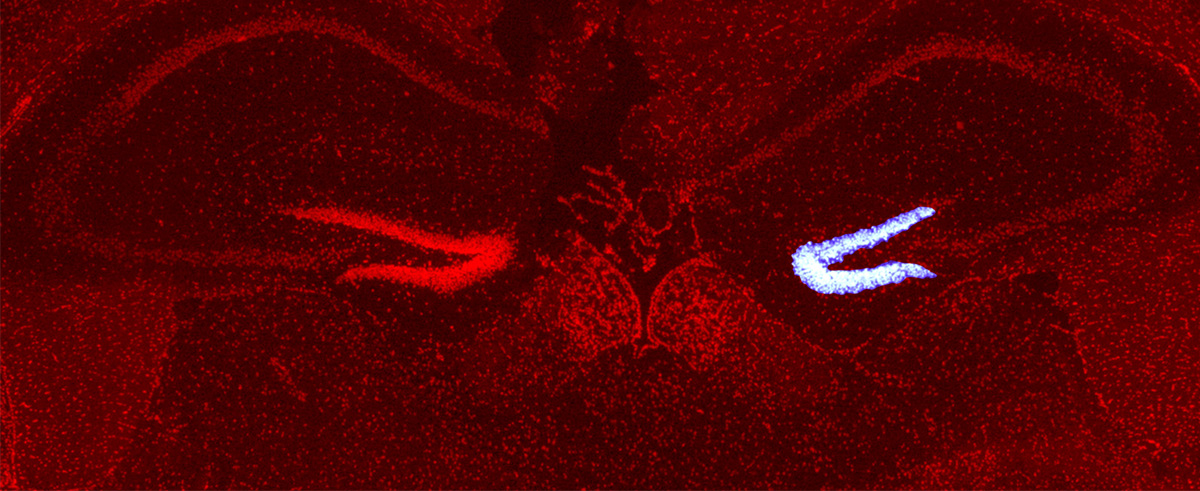
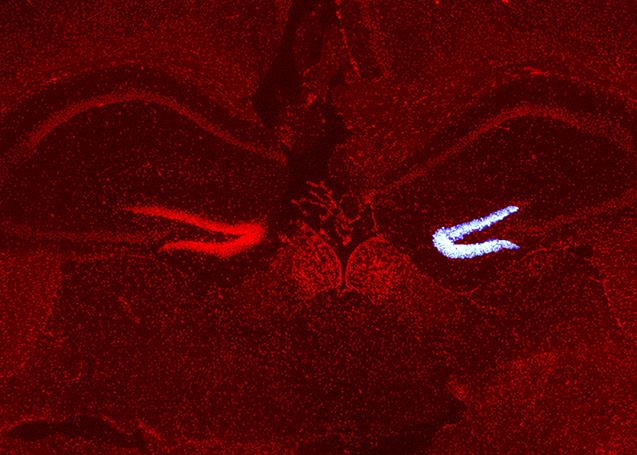
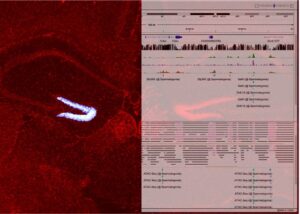
- 2025-01-07名古屋市立大学の澤本和延先生らのグループとの共同研究論文が Frontiers in Neuroscience 誌に公開されました。本田瑞季さんがマウス脳の損傷部位に移動中の神経芽細胞に対する PIC RNA-seq を実施し、TGFβ シグナルが更新していることを見出しました。
- 2024-12-06東京大学の加藤大志先生らのグループとの共同研究論文が Science Advances 誌に公開されました。木村龍一さんがムンプスウイルス感染で生じる封入体に対する PIC RNA-seq を実施し、G4 構造を持つ RNA が濃縮していることを見出しました。本論文は東大と熊大の共同でプレスリリースしました。
- 2024-12-03鄒 兆南さんが東京理科大学創域理工学部で ChIP-Atlas ハンズオンセミナーを開催しました。
- 2024-11-27理研の粕川雄也先生らのグループとの共同研究論文が Nucleic Acids Research 誌に公開されました。沖 真弥さんが開発した ChIP-Atlas と、粕川先生が開発した fanta.bio との連携が記載されています。
- 2024-11-25沖 真弥さんが大阪大学超実践的バイオインフォマティクスセミナーで講演しました。
- 2024-10-23鄒 兆南さん、沖 真弥さんが APBJC2024 にて講演しました。